|
|
Leadership and Team membersChair:
Macquarie University, Sydney, Australia Co-Chairs:
Daniel Kolarich, PhD Griffith University, Gold Coast, Australia
Nicolle H Packer, PhD Macquarie University, Sydney, Australia Early Career Researchers:
Rebeca Kawahara, PhD Macquarie University, Sydney, Australia
Tiago Oliveira, PhD IMBA, Austrian Academy of Sciences, Vienna, Austria |
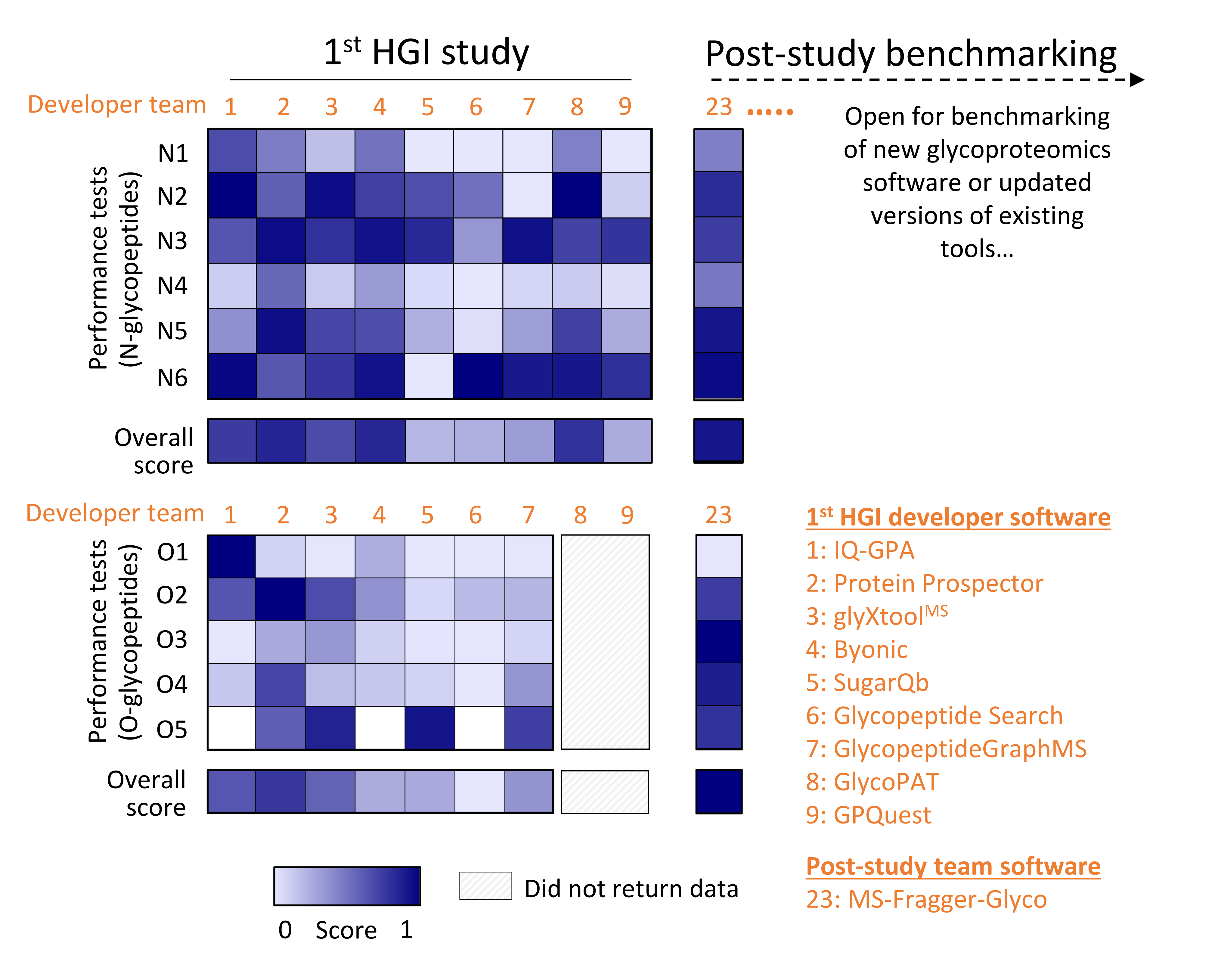
Post-study benchmarking of recent glycopeptide software against developers included in the 1st HGI study To make best use of methodology and outcomes of the 1st HGI study and to bridge to the 2nd HGI study, which is well underway, the Human Glycoproteomics Initiative are here making the community aware of a post-study benchmarking opportunity for new and updated software against the performance of the nine developer teams that completed the 1st HGI study (Kawahara et al., Nat Methods, 2021). The accuracy and value of such a post-study benchmarking effort is critically dependent on the adherence to the published study guidelines. Notably, the resulting performance profiles should be viewed in the light that software included in the post-study phase may have benefitted from completing the study after the publication of the 1st HGI study outcomes. Developers interested in the benchmarking of new and updated versions of existing software for glycopeptide data analysis can contact A/Prof Morten Andersen or Dr Rebeca Kawahara to discuss further. The performance profiles of the post-study benchmarking will be made publicly available through this site to benefit the community. The first software to be tested in this post-study benchmarking effort was MSFragger-Glyco (Polasky et al., Nat Methods, 2020), which missed the 1st HGI comparison due to the development of the software after the study evaluation period. The MSFragger-Glyco developer team headed by Prof Nesvizhskii and Dr Dan Polasky completed the post-study in April 2022 using a new release of MSFragger-Glyco version v3.5 in FragPipe 18.0 according to the study guidelines and submitted their lists of identified N- and O-glycopeptides using the common HGI reporting template to HGI-affiliated Dr Rebeca Kawahara who analysed the data in May 2022. Dr Kawahara established the relative performance profile of MSFragger-Glyco using the same scorecard system as used in the 1st HGI study. The performance profile of MSFragger-Glyco relative to the performance of the original nine developer software solutions suggests that MSFragger-Glyco is a promising high-performance tool not least for O-glycoproteomics where this search engine stood out from other tested software (see associated Figure). The Human Glycoproteomics Initiative acknowledges Dr Rebeca Kawahara for performing the post-study benchmarking of MSFragger-Glyco and Prof Alexey Nesvizhskii and Dr Dan Polasky for completing the glycopeptide challenge and for agreeing to making the performance data of MSFragger-Glyco publicly available.
Other Community Initiatives in the Glycosciences:The Human Glycome Project | Announcements & upcoming events2nd HGI Study now open and recruiting glycoproteomic software developer teams Deadline to join is May 1, 2022 27th Annual Lorne Proteomics Symposium 2022 Saturday, Feb 5 - 4:00PM Community Evaluation of Glycoproteomics Software - Morten Thaysen-Andersen
Future HGI studies Follow-up efforts seeking to compare the performance of the most recent glycoproteomics software upgrades and informatics solutions not included in the 1st HGI study are being drafted within the next study of the Human Glycoproteomics Initiative. Studies addressing key challenges relating to glycopeptide data acquisition are also planned. Community-wide calls for participation in these studies will be made soon.Further ReadingRecent glycoproteomics reviews, non-exhaustive list... |







.png)