|
|
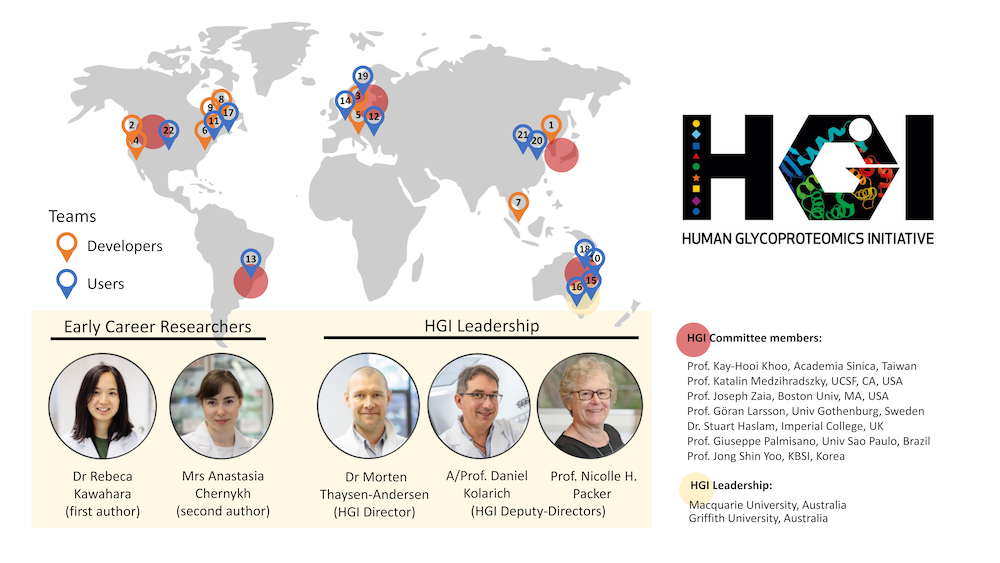
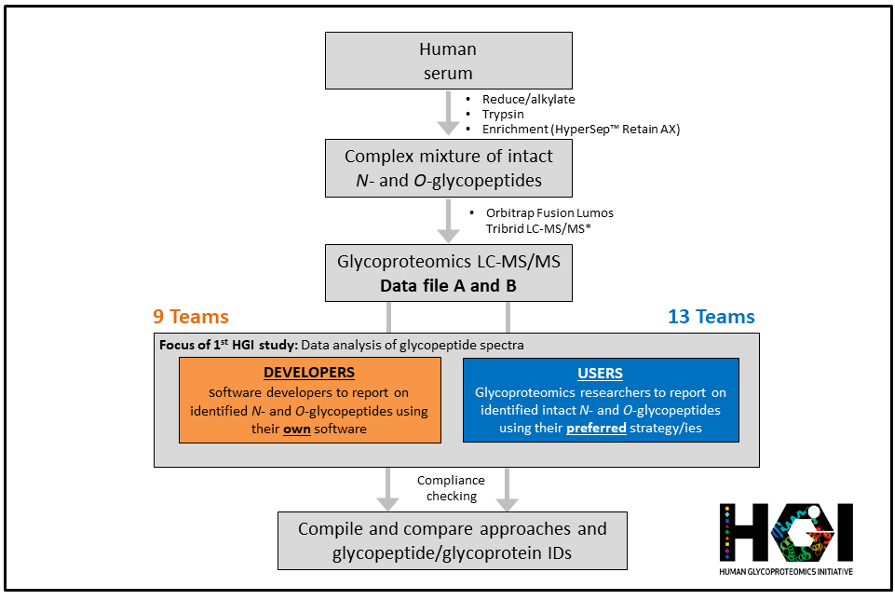
“A goal of the 1st Human Glycoproteomics Initiative (HGI) Study was to assess the relative performance of current glycoproteomics software for intact N- and O-glycopeptide identification from high resolution MS/MS spectral data across laboratories.” Profiling of intact glycopeptides at scale from complex biological mixtures (glycoproteomics) remains a considerable analytical challenge and a frontier in proteomics. It is recognised that the lack of efficient search engines tailored to the unique challenges associated with large-scale glycopeptide analysis continues to hinder the rapid expansion and democratisation of glycoproteomics technologies beyond specialist laboratories. Excitingly, the field has seen the development of many innovative bioinformatic tools over the past decade that promises to streamline and semi-automate the glycopeptide identification process based on high content tandem mass spectrometry data. Conducted through the HUPO B/D-HPP – Human Glycoproteomics Initiative (HGI), this community-driven 1st HGI study brought together field-leading developers and expert users of glycoproteomics software to evaluate the performance of informatics solutions for system-wide glycopeptide mass spectrometric analysis [1]. In total, 25 teams from 11 countries across five continents signed up for the challenge out of which 22 teams (~90%) completed the study. The study design including the sample type, preparation and data collection method was carefully chosen to mimic conditions typically encountered in glycoproteomics analysis while also aiming to accommodate most available informatic solutions and to appeal to users in the field. All teams were asked to identify intact N- and O-glycopeptides from two shared high-resolution LC-MS/MS data files (Data file A and B) of N- and O-glycopeptides from human serum proteins (LC-MS/MS data kindly provided by Drs Rosa Viner and Sergei Snovida, Thermo Fisher Scientific) and report back their identifications and their search strategies in a comprehensive and standardised reporting template. Completed reports were thoroughly checked by the study organisers for compliance to the study guidelines to enable a fair comparison between teams.
In short, we conclude from this study that diverse software for comprehensive glycopeptide data analysis exist, point to several high-performance search strategies, and specify key variables that may guide future software developments and assist informatics decision-making in glycoproteomics data analysis. While informatics challenges undoubtedly still exist in glycoproteomics, our study interestingly highlights that several computational tools, some already demonstrating high performance, others considerable potential, are available to the community. 1st HGI Study committee: Dr. Morten Thaysen-Andersen, Macquarie University, Australia (Chair) Prof Nicki Packer, Macquarie University and Griffith University, Australia A/Prof. Daniel Kolarich , Griffith University, Australia | Announcements & upcoming eventsFuture HGI studies Follow-up efforts seeking to compare the performance of the most recent glycoproteomics software upgrades and informatics solutions not included in the 1st HGI study are being drafted within the next study of the Human Glycoproteomics Initiative. Studies addressing key challenges relating to glycopeptide data acquisition are also planned. Community-wide calls for participation in these studies will be made soon. |