Sri H. Ramarathinam and Anthony W. Purcell, Department of Biochemistry and Molecular Biology and Infection and Immunity Program, Biomedicine Discovery Institute, Monash University, Clayton 3800, Victoria, Australia
#immunosurveillance
Our immune system sources actionable intelligence in the fight against pathogens in many forms including peptides, lipids and other small molecules associated with infection and cancer. The Human Leukocyte Antigen (HLA) proteins, expressed on cell surface, play an important role in conveying the status of cellular health to the immune system by presenting short peptides to T-cells. These short peptides could be from a variety of cellular and extraneous sources forming a snapshot of proteins - synthesized or degraded within the cell. Two major pathways enable antigen presentation: the HLA class I, expressed on all nuclear cells, present endogenous antigen to CD8+ T cells and the HLA class II, found only on professional antigen-presenting cells (such as dendritic cells and macrophages) present antigen from endogenous and exogenous sources to CD4+ T cells (Figure 1).
Specific receptors on T-cells (TCRs) are used to survey the landscape of HLA-peptide ligands on cell surface to find their cognate peptide-HLA complex, much like going through social media feeds to find a specific post of interest. Each TCR can generally recognise a single HLA-peptide complex, due to the remarkable sequence diversity engendered in the TCR through recombination of different genetic elements during T-cell development. T-cells also undergo selection in the thymus such that they are poised to recognise foreign peptides that may arise from viral and bacterial proteins or from inappropriately expressed or somatic mutation-bearing peptides in cancer. Presentation of such foreign peptides in complex with HLA molecules on the surface of infected or malignant cells attracts scrutiny of T-cells bearing the TCR that can recognise this HLA-peptide complex resulting in activation of the immune response and eradication of the cell. The ‘status updates’ in form of immune-related peptides constitutes the ‘immunopeptidome’ of the cells and has been characterised in multiple host species including human, mouse, bat [1], bovine [2], swine [3] and chicken [4].
#immunopeptidomics
The term immunopeptidomics describes the systematic, high-throughput analysis of HLA-bound peptides using mass spectrometry [5-7]. Understanding and deciphering the cellular communication updates (peptide sequences) by the immune system is crucial in the development of new vaccines against viruses as well as immunotherapies against cancer and auto-immune diseases [8-10].
#challenges
Identifying HLA-peptides involves immunoprecipitation of HLA molecules from cells or tissues followed by separation of peptides from heavy chains, fractionation and subsequent analysis by mass spectrometers [11]. In contrast to global proteomic analysis, the HLA peptides pose unique challenges requiring exacting sample preparation and analysis strategies. There is a need to have dedicated and specialised labs and informatics pipelines to overcome some common challenges. The yield depends on expression of HLA molecules, choice of appropriate methodologies to isolate the peptides, fine tuning of parameters for acquisition of high-quality mass spectrometry data and ultimately appropriate software to interpret data and identify peptide sequences.
One of the biggest hurdles is the amount of material required to do an in-depth immunopeptidomics analysis. To overcome the sample limitation, several algorithms have been developed to predict peptides that bind to specific HLA [12, 13]. While the prediction tools are invaluable, they do not yet consider post-translationally modified (PTM) peptides. Additionally, there is a disparity between peptides identified by mass spectrometry and the top results from prediction algorithms with the majority only explaining at most 10% of peptides as strong and or weak binders. Developments in sample processing and sensitive instrumentation are reducing the need to have large sample sizes and several studies already analysing patient material either individually or in small pools. The HLA-peptides bearing diverse C-termini, for example, may require analysis of singly-charged ions which are traditionally ignored in proteomics workflows [14].
The software to search the MS data is another area that has contributed significantly to improve the number of peptides identified. There is scope for improvement, especially development of appropriate decoy databases for HLA peptides that tend to have a variety of N- and C-termini and in developing peptide-centric algorithms without any influence of protein grouping. Endogenous processing in cells and HLA type result in varied peptide, requiring significant computing resources during database search.
A community standard of cell lines that can be used to benchmark the complete process (cell line to data) will also be an invaluable tool to improve upon and push the limits of current techniques. Additionally, there is a need for robust well-defined, widely-available synthetic HLA-peptide standards (>1000-5000 peptides) to benchmark various peptide identification and informatics pipelines.
#HUPO-HIPP
Formation of HUPO-Human Immunopeptidome Project (HUPO-HIPP) in 2015, brought the leaders in the field together to advance research, support collaborative efforts including development of standards for publication [15, 16]. HUPO-HIPP organised two successful events so far, including a summer school and a precision oncology meeting that introduced the techniques and highlighted some key challenges in the path forward. New members are welcome to join us at HUPO-HIPP for latest updates.
In recent times, there is a growing appreciation for presence of PTM peptides [17] and peptides that are non-genomically templated in addition to the linear peptide sequences. While they challenge known dogmas, peptides from Defective Ribosomal Products (DRIPs) [18], post-translationally spliced peptides [19, 20] and peptides from UTRs and non-coding regions are worthy of consideration to explore their potential role in health and disease. As a field, tremendous achievements have led to a deeper understanding of the antigen presentation and processing machinery, yet we continue to be surprised by the intricate network of cells and proteins that keep us safe
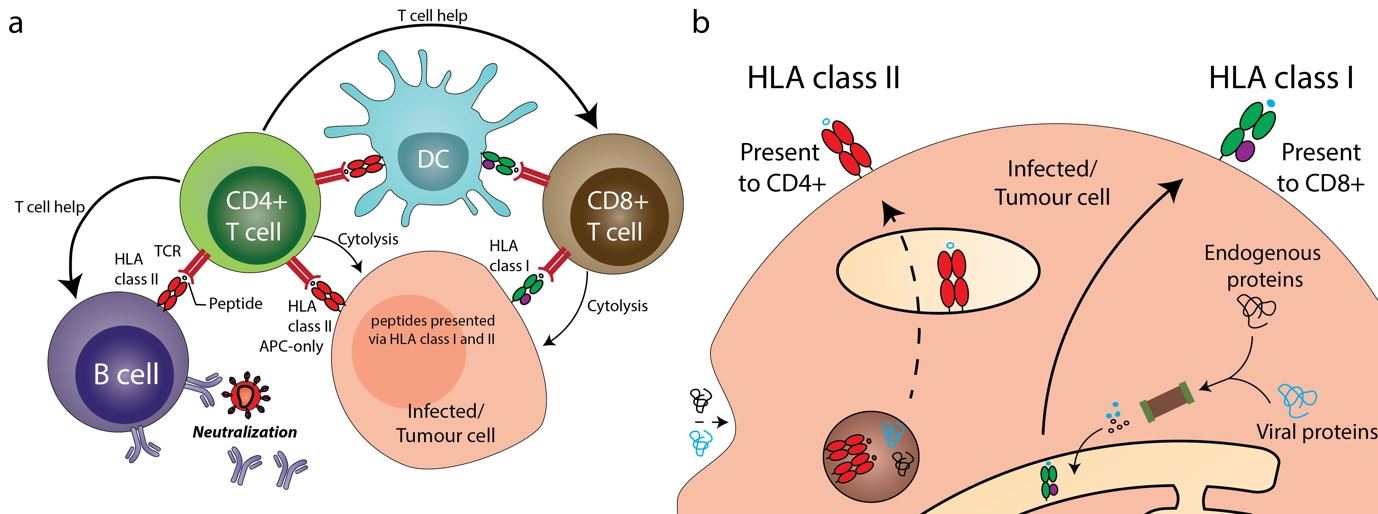
Figure 1: A) HLA-class I and II molecules play a crucial role in key immunological pathways. Understanding the peptide repertoire offers insight into immunosurveillance machinery and its modulation. B) HLA class I and II pathways present antigen to CD8+ and CD4+ T-cells respectively.
References:
1. Wynne, J.W., et al., Characterization of the Antigen Processing Machinery and Endogenous Peptide Presentation of a Bat MHC Class I Molecule. J Immunol, 2016. 196(11): p. 4468-76.
2. Nielsen, M., T. Connelley, and N. Ternette, Improved Prediction of Bovine Leucocyte Antigens (BoLA) Presented Ligands by Use of Mass-Spectrometry-Determined Ligand and in Vitro Binding Data. Journal of Proteome Research, 2018. 17(1): p. 559-567.
3. Pedersen, L.E., et al., Porcine major histocompatibility complex (MHC) class I molecules and analysis of their peptide-binding specificities. Immunogenetics, 2011. 63(12): p. 821-834.
4. Cumberbatch, J.A., et al., Chicken major histocompatibility complex class II molecules of the B19 haplotype present self and foreign peptides. Animal Genetics, 2006. 37(4): p. 393-396.
5. Caron, E., et al., The structure and location of SIMP/STT3B account for its prominent imprint on the MHC I immunopeptidome. Int Immunol, 2005. 17(12): p. 1583-96.
6. Hunt, D.F., et al., Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science, 1992. 255(5049): p. 1261.
7. Falk, K., et al., Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature, 1991. 351(6324): p. 290-296.
8. Engelhard, V.H., et al., MHC-restricted phosphopeptide antigens: preclinical validation and first-in-humans clinical trial in participants with high-risk melanoma. J Immunother Cancer, 2020. 8(1).
9. He, Q., et al., Targeting cancers through TCR-peptide/MHC interactions. Journal of Hematology & Oncology, 2019. 12(1): p. 139.
10. Serra, P. and P. Santamaria, Antigen-specific therapeutic approaches for autoimmunity. Nature Biotechnology, 2019. 37(3): p. 238-251.
11. Purcell, A.W., S.H. Ramarathinam, and N. Ternette, Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc, 2019. 14(6): p. 1687-1707.
12. Jurtz, V., et al., NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol, 2017. 199(9): p. 3360-3368.
13. Peters, B., M. Nielsen, and A. Sette, T Cell Epitope Predictions. Annual Review of Immunology, 2020. 38(1): p. 123-145.
14. Pandey, K., et al., In-depth mining of the immunopeptidome of an acute myeloid leukemia cell line using complementary ligand enrichment and data acquisition strategies. Mol Immunol, 2020. 123: p. 7-17.
15. Lill, J.R., et al., Minimal Information About an Immuno-Peptidomics Experiment (MIAIPE). Proteomics, 2018. 18(12): p. e1800110.
16. Admon, A. and M. Bassani-Sternberg, The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol Cell Proteomics, 2011. 10(10): p. O111.011833.
17. Mei, S., et al., Immunopeptidomic analysis reveals that deamidated HLA-bound peptides arise predominantly from deglycosylated precursors. Mol Cell Proteomics, 2020.
18. Wei, J. and J.W. Yewdell, Flu DRiPs in MHC Class I Immunosurveillance. Virol Sin, 2019. 34(2): p. 162-167.
19. Faridi, P., et al., A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci Immunol, 2018. 3(28).
20. Liepe, J., et al., A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science, 2016. 354(6310): p. 354-358.


.png)